In 1986, the nervous system of Caenorhabditis elegans, a microscopic worm, was fully mapped. At the time, scientists and engineers thought this map would quickly reveal the definite functions of the nervous system. But it turned out that resolving the functions of a nervous system is very complex and even today, researchers still don’t fully understand how neural structure transforms to function.
But recently, the Department of Electrical & Computer Engineering and Applied Mathematics Washington Research Foundation assistant professor Eli Shlizerman and ECE graduate student Jimin Kim, created an interactive simulation of the nervous system of the worm, called the Neural Interactome. The purpose of their open source creation, is to help others better understand the neuronal system.
“We designed this interactive tool for exploring both the architecture and the dynamic functions of neurons at the same time,” said Shlizerman. “Our web platform can be stimulated to see in real-time how neurons’ dynamics change.”
Their research, “Neural Interactome: Interactive Simulation of a Neuronal System,” was published in “Frontiers in Computational Neuroscience,” in March.
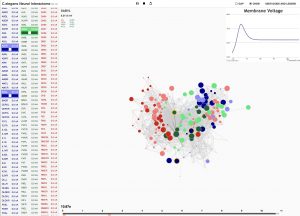
Interactive interface for Neural Interactome. Left panel enlists all the neurons classified by type (sensory, inter and motor). Each neuron is a clickable button with a scroll option. Scrolling adjusts the magnitude of constant stimulus; shift + click ablates the neuron from the network. Right: Force-directed graph displays each neuron’s membrane voltage (node color denotes the sign; radius denotes the magnitude) and connections between neurons (edges between each pair of nodes).
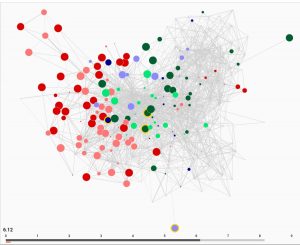
The cross-platform framework that the pair built, which combines graph visualization with the simulation of neural dynamics, shows the many connections and how these support interactions between neurons. On the web platform, a graph represents the full somatic nervous system where each circle is a neuron and the different colors showcase the different classes of neurons: sensory, motor and interneurons. Users can hover their mouse over each neuron and the Neural Interactome will show them where it is in the nervous system, as well as the chemical and electrical connection it has.
“You can see all of the connections and possible interactions between the neurons,” Kim said. “When you choose stimuli and hit play, the circles representing the neurons change in size and color expressing their membrane voltage. Effectively, we see how the full nervous system comes to life and responds to stimuli in real time. You can see how the neurons are communicating which composes the “thinking” of the worm.”
Because the simulation is open-sourced, anyone with interest in the neural system can see how it performs. Click on a neuron, disable it, and see how the system’s dynamics change in real time.
“There are still a lot of neurons which scientists haven’t figured out their purpose, even in the Caenorhabditis elegans,” Shlizerman said. “People thought that by the 90s, researchers would resolve most of the functions of this tiny neural system, but experiments only showed how complex it is, and how much there is to learn.”
The interactome demonstrates that even the effect of disconnecting a single neuron, such that it is not functioning, changes the way of those that are, work.
Shlizerman said that in 1986, John Graham White, a professor of anatomy and molecular biology at the University of Wisconsin-Madison, reconstructed the nervous system of the Caenorhabditis elegans, under the supervision of Nobel Laureate Sydney Brenner. This turned out to be a start for simulating all of the neurons in the nervous system.
“Thirty years later, we continued the work and created this tool which simulates the dynamics and connections,” Shlizerman said. “We also developed visualization methods to highlight neurons belonging to the same functional groups so it will be easier to cluster them when they are performing their function.”
Kim said the latest data is organized using the common Python 3 platform to simulate the network, and the hope is that computational scientists will keep revising the data and work with the Interactome to add more layers to the network so everyone can see their effects.
Researchers believe that their work could be applied to other more complex nervous systems, such as the brain of the fly or the mouse. Shlizerman said that brain mappings, called connectomes, for these organisms are currently being rapidly resolved and they include much more neurons than the 302 neurons of Caenorhabditis elegans.
“As mappings of those organisms become more detailed, our framework paves the way to study how neural interaction and structure generate behavior,” he said.
The project was sponsored by the National Science Foundation and Washington Research Foundation. Shlizerman is also a fellow at the UW eScience Institute and on the leadership team of the UW Computational Neuroscience Center.